Exploring genomic methods for delimiting species in radiations of terrestrial snails
Delimiting species in radiations is notoriously difficult because of the small differences between the incipient species, the star-like tree with short branches between species, incomplete lineage sorting, and the possibility of introgression between several of the incipient species. Next generation sequencing data may help to overcome some of these problems. We evaluated methods for species delimitation based on genome-wide markers. For this purpose, we developed in cooperation with Prof. Christian Hennig from the University of Bologna a novel approach for testing isolation-by-distance, i.e. the hypothesis that the multilocus genetic distances between individuals or populations belonging to two different candidate species are not larger than expected based on their geographical distances and the relationship of genetic and geographical distances within the candidate species (implemented in the R package Prabclus). A rejection of this null hypothesis is an argument for classifying the two studied candidate species as distinct species.
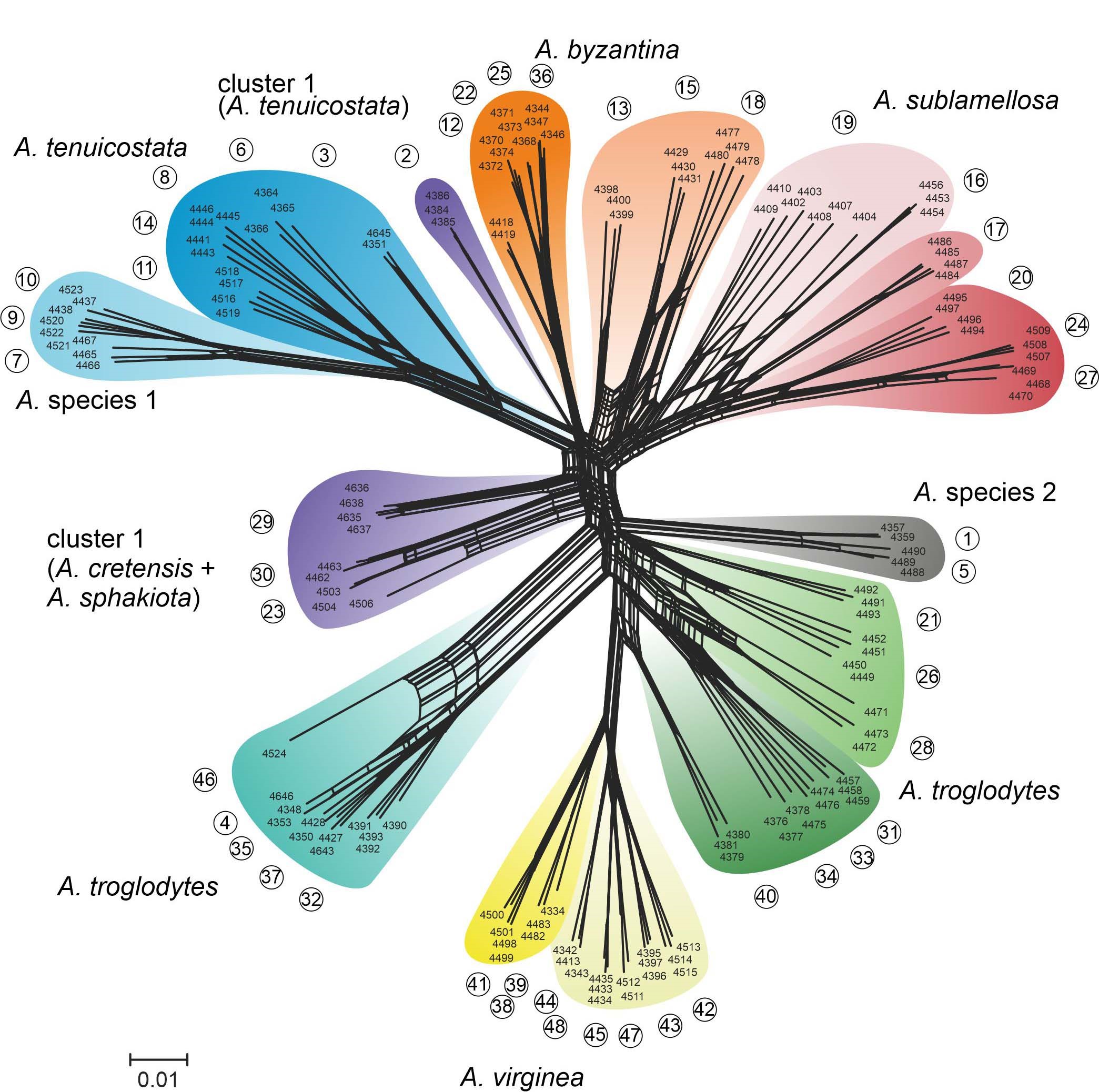
In the first phase of the project, we used double-digest restriction site-associated DNA (ddRAD) sequencing data of the radiation of the Albinaria cretensis group on Crete to evaluated methods for delimiting species. Previously species delimitation in the Albinaria cretensis group was based exclusively on shell characters and resulted in classifications distinguishing 3–9 species. Based on ddRAD data of 48 populations of the Albinaria cretensis group, we evaluated three methods for species discovery. The multispecies coalescent approach implemented in the program Bayesian Phylogenetics and Phylogeography resulted in a drastic overestimating of the number of species, whereas Gaussian clustering resulted in an overlumping. Primary species hypotheses based on the maximum percentage of the genome of the individuals derived from ancestral populations as estimated with the program ADMIXTURE moderately overestimated the number of species, but this was the only approach that provided information about gene flow between groups. Two of the methods for species validation that we applied, BFD* and delimitR, resulted in an acceptance of almost all primary species hypotheses, even such based on arbitrary subdivisions of hypotheses based on ADMIXTURE. In contrast, secondary species hypotheses, resulting from an evaluation of primary species hypotheses based on ADMIXTURE with isolation-by-distance tests, approached the morphological classification, but also uncovered cryptic species and indicated that some of the previously delimited units should be combined. Thus, we recommend this combination of approaches that provided more detailed insights in the distinctness of barriers between the taxa of a species complex and the spatial distribution of admixture between them than the other methods.
In the next phase of the project, we will apply the developed approaches to resolve the taxonomy of land snail radiations in Indonesia and to understand the causes of these radiations. Indonesia covers two of the global biodiversity hotspots, Sundaland and Wallacea and its fauna. This project will be done in cooperation with Dr. Ayu Nurinsiyah from the Zoological Museum of the Indonesian Institute of Science in Java. Previous studies of the land snail fauna of Java together with Dr. Ayu Nurinsiyah have shown that a high number of species has still to be discovered and described.
Doctoral thesis Sonja Bamberger (funded by the DFG priority program 1991 Taxon-Omics: New approaches for discovering and naming biodiversity).
- Hausdorf, B. & Hennig, C. 2020. Species delimitation and geography. Mol. Ecol. Resour., 20: 950–960.
- Nurinsiyah, A. S., Neiber, M. T. & Hausdorf, B. 2019. Systematic revision of the genus Landouria Godwin-Austen, 1918 (Gastropoda, Camaenidae) from Java. Eur. J. Taxonomy, 526: 1–73.